Research Software Directories
This is a summary of a SORSE discussion session, presented by:
- Mark Woodbridge, Imperial College London
- Vanessa Sochat, Stanford University
- Jurriaan Spaaks, Netherlands eScience Center
And featuring contributions from:
- Malin Sandström, INCF
- Alexander Struck, Humboldt University of Berlin
Introduction
The discussion session “Research Software Directories: What, Why, and How?” was held on September 16 during SORSE, an International Series of Online Research Software Events. As presenters, we each shared efforts to develop and maintain software directories: catalogues to showcase the software outputs of an institution or community. The directories presented were:
- The Research Software Directory developed by the Netherlands eScience Center (GitHub)
- Imperial College London’s Research Software Directory (GitHub)
- The Research Software Encyclopedia
- GitHub Search as a template for an individual or institution
Each of the above offered several advantages and disadvantages, or were scoped for particular use cases. For example, research-software.nl provides a robust application for serving detailed metrics and metadata for software, however it requires more manual entry. The Research Software Encyclopedia is automated and does not require hosting, but it lacks the same level of metadata. The Imperial College London and GitHub Search research software directories offer much quicker to deploy solutions, but might be too simple for some use cases. The directories are discussed in detail in the following sections. In addition to this set, we suggest the reader take a look at the Awesome Registries list to find additional examples.
How many participants use software directories?
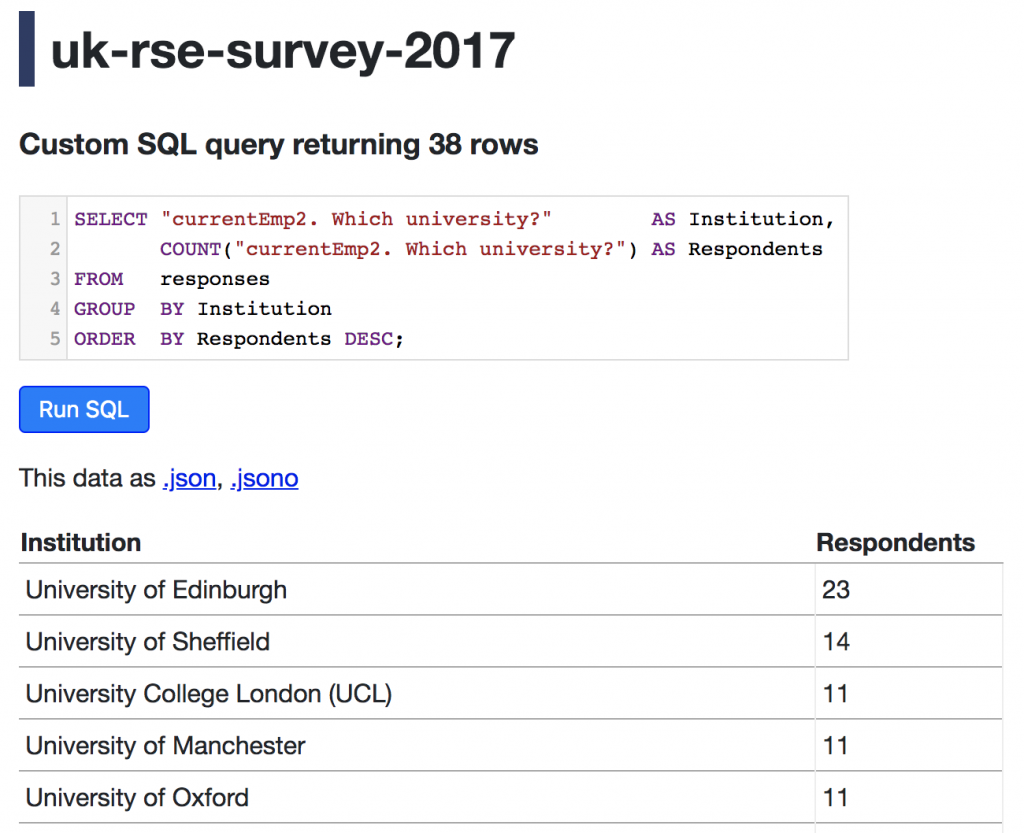
We were quite surprised at the results of asking attendees the extent to which they have contributed or used software directories. For a total of 27 participants, 43% have used a directory for a relevant project, 27% have submitted software to a directory, and 58% indicated neither of the above.
Presentations
The Research Software Directory by Netherlands eScience Center
- Slides available from Zenodo: doi: 10.5281/zenodo.4034290
- YouTube video (starting 3:20)
Jurriaan’s presentation started off by explaining why the Netherlands eScience Center had a need for what eventually became the Research Software Directory. Primary reasons were that as the Netherlands eScience Center grew beyond say, 20 or so engineers, tracking what software was available in-house really became too difficult a problem to do ad-hoc, despite the fact that all of their repositories are publicly accessible on GitHub. Secondly, the eScience Center strives to be as open as possible, and they thought it was important to be able to show the outside world where the taxpayer’s money had gone. Lastly, the eScience Center has a continuous need to keep track of various metrics, both for reporting to their funders (SURF and NWO), but also for helping management make informed business decisions.
Jurriaan then demonstrated the eScience Center’s instance of the Research Software Directory. While walking the viewers through the design, he explained how the product pages’ design was helping site visitors on their way towards adoption of the software presented on the product page.
When designing the Research Software Directory, specific attention was paid to how an instance is filled with data, how this data is curated, and how to do this in a way that can be sustained over time. To this end, the Research Software Directory harvests much of its information automatically, for example using APIs to GitHub (code development platform), Zenodo (archiving service), and Zotero (reference manager). This setup allows engineers employed by the Netherlands eScience Center to stay mostly in their comfort zone (i.e. GitHub). They just need to make sure to follow best practices such as having publicly accessible repositories, making releases on Zenodo using the automated integration, and including software citation metadata (CFF) in their repositories. Given that they already do much of that anyway, making an entry in the Research Software Directory can be achieved in a few clicks via the Admin interface that the Research Software Directory provides.
The Research Software Directory has proven to be a great resource for managing the organization, for providing funders with relevant metrics, and for increasing the visibility of tools. Despite these upsides, of course there are some downsides as well, for example it has proven difficult to carve out enough time to curate prose on the product pages, leading to text snippets that are sometimes too difficult to read for visitors not yet familiar with the software that the product page presents. A second problem is maintenance of the Research Software Directory software itself: the software stack includes more than 40 techniques, methods, and tools, in various languages and using a variety of frameworks. It has proven difficult to find developers that are familiar enough with all of these to be effective at maintaining the site. While this has not led to any significant downtime in the 3 years research-software.nl has been running, eScience Center intends to start reducing the software stack in the very near future. Furthermore, they are investigating whether it’s feasible to provide Research Software Directories as a service.
The Research Software Directory by Imperial College London
- Slides available from Zenodo: 10.5281/zenodo.4058207
- YouTube video (starting 20:01)
Mark Woodbridge demonstrated Imperial College’s Research Software Directory, explaining how it was developed to present a manually curated list of GitHub and GitLab repositories – motivated by a desire to rapidly catalogue and demonstrate the breadth of software developed at Imperial. It is also intended to encourage collaboration by assisting researchers to identify existing expertise and projects at Imperial.
The chosen approach has resulted in a system which is easy to maintain – both in operational complexity and in adding entries to the directory (even if the latter does depend on some familiarity with git and GitHub i.e. making a commit and pull request). This simplicity comes at a price: it depends on Algolia (a freemium service), has limited features, and is not easy to customise. It also relies on manual curation and repository metadata: due to limited bandwidth and lack of incentives, developers rarely submit or annotate software themselves. Finally, it lacks the polish and level of detail that you might expect of a public-facing showcase.
The system has however achieved its aims in effectively showcasing research software and developers at Imperial, and has provided a set of metadata enabling the identification of preferred languages to fast-growing fields of research. A suite of standalone utility scripts ensures that the contact details and project web pages remain up-to-date, and that new repositories by known developers are added to the directory in a timely manner.
The Research Software Encyclopedia
- Slides available from Google Slides
- YouTube video part 1 (starting 30:26)
- YouTube video part 2 (starting 45:25)
The Research Software Encyclopedia (RSEPedia) is a community-driven, open source directory that provides a means to communicate about software. It consists of three components – a set of criteria and taxonomy items used to describe or otherwise communicate about software categorization preferences, a database, and a command line client to interact with those components. The criteria and taxonomy items are maintained in their own GitHub repository, https://github.com/rseng/rseng, and render to an interface to allow for exploration and visualization. Importantly, the site for these items hosts a weekly software showcase, allowing the community to learn more about open source libraries that might be useful for their work. The terms are also served programmatically to a RESTful application programming interface (API) that makes them readily available for the RSEPedia software, which is also provided on GitHub (https://github.com/rseng/rse). Using the software, an individual or institution is empowered to easily generate a database and interface for a set of software they care about. They can inspect, add, search, or otherwise interact with metadata. While relational databases can be created, the community maintained database is a flat file database hosted on GitHub (https://rseng.github.io/software) that allows an interested contributor to browse, and annotate software with criteria and taxonomy items in an online interface. Annotation only takes a few clicks, and the process to make changes and update the database is fully automated via GitHub actions. Annotation in bulk is also easy to do locally after cloning the software repository, starting the annotation interface, and opening a pull request with changes. Importantly, although annotation can help to share ideas about software, it is not required to make the RSEPedia useful. By way of being able to communicate about software via asking questions, and by way of the software showcase, the RSEPedia can be successful for your needs if you just need a way to describe what you are looking for (e.g., for a grant or journal) or just want to share your set of software to be easily searchable.
GitHub Search is a derivation of the Research Software Directory by Imperial College London, but it removes the Algolia dependency, and derives software repositories directly from the GitHub API list of repositories for an organization directly on GitHub pages. This means that deployment is easy, coming down to simply creating the repository with a GitHub action to build it at some frequency to update the pages.
Discussions
After the presentations, attendees were divided over three groups for a 20-minute discussion session. All three groups saw lively discussions and discussed a plethora of relevant subjects, a selection of which is included below.
How do software directories interact with high performance computing (HPC)?
With several attendees that work as administrators for HPC, the question quickly came up about the relationship between software directories and HPC centers. Indeed, these centers typically maintain a large catalog of software for a user base, and it could be beneficial to link this software catalog or strategy to maintain it with a software directory. For example, if you are familiar with spack or easybuild you could imagine having integration to use a software directory to look up metadata, or generate user-friendly documentation pages. The pages might have install instructions, examples, and optimization hints for different architectures.
Guix-HPC is a package manager for a variety of software that is developed to allow reproducible HPC environments. It may interact with existing instances of Research Software Directories.
Curation policies
The main concern related to the “curation” of software directories were criteria for inclusion. A lively discussion related to the definition of “research software”, particularly in relation to scale and licensing. In the broadest sense there was agreement in principle that it could refer to any tool or library used to produce scientific results.
In terms of scale, attendees working in life sciences research emphasized that research software in their context could be a standalone script, and software directories should therefore “scale-down” appropriately. Scripts of this type may be less substantial but their quality could well be assessed similarly to more prototypical projects in terms of documentation, design for re-use and version control.
Licensing was a more challenging topic – an argument was made for directories enabling users to find any tool that might accelerate research, including commercial software – as long as an appropriate licence was available.
In broader terms, there was consensus that curators should avoid making assumptions about software applicability and relevance, even if they do have domain knowledge. More important than strict policies is effective annotation and filters so that users can apply their own criteria when searching for relevant software.
Searching for software
Searching for software presents its own challenges as an RSD only presents local results and many other platforms would need to be consulted for an exhaustive overview of relevant packages. Here, some registry lists prove to be helpful, for example Awesome Research Software Registries.
The purpose and minimum features of Research Software Directories
Participants identified discoverability as a major issue in relation to research software, particularly for domain specialists (i.e. end-users). This led to the following features being considered of primary importance:
- Metadata clearly explaining the purpose and value of individual software tools in non-technical terms. The community is currently working on metadata standards like CFF or CodeMeta.
- Contact details for the authors of the software in case further advice or support is required
- Installation and getting started instructions
- Guidance on how to cite the software
- Licensing terms. This was discussed not only in relation to terms of use but also, for non-free software, ensuring cost-efficiencies by avoiding unilateral purchasing decisions and promoting the use or procurement of shared/group licences.
Many other features may benefit researchers, for example, linking from an RSD entry to its accompanying paper and data, as suggested in “Generalist Repository Comparison Chart” or listing received software citations, as implemented in swMATH.
Organization-based registry vs community-based registry
Some registries out there are scoped to serve an organization, whereas other registries like ascl.net or bio.tools aim to serve an entire research community. An advantage of the latter is increased traffic to the registry, and real benefits for users to browse the registry to see if somebody else in the community already created a solution. However, because the social structure across the community is quite loose, it will be more difficult to keep people involved, to discover new tools that could be added to the registry, and to make sure that the language used on the registry’s pages is understandable by everyone in the community. Furthermore, governance of the instance will be more difficult. For example, within the community there may exist different opinions on what metadata should be kept, and weighing these opinions will be more difficult in a larger community than a small one.
In contrast, organizational registries are more easy to run and govern — discovering tools that could be added is (or used to be) a matter of hanging out at the coffee machine and asking your colleague what they are working on right now. Helping your colleague enter their data, and making sure they do it correctly, is easier as well, and some good old-fashioned peer pressure can be applied if needed. Funding policies currently do not mandate the publication of research software, as Horizon 2020 required for research data (if possible).
Further resources
- A categorization by type of information or content in the registry is provided by “Research Software Discovery: Challenges, Risks and Opportunities“.
- Re3Data provides an overview of registries, including both research software and data
- The Research Organization Registry provides an overview of Research Organizations
Recommendations and Next Steps
By discussing topics of curation, federation, technology and sustainability of research software directories with a wider audience, this discussion section hoped to not only promote the benefits of such directories and encourage their deployment, but also to identify issues and gather ideas to address them. From discussion above, it’s clear that there are interesting projects and updates to existing directories that might be pursued.




 In this post we’ll demonstrate how to run deploy a simple scheduled task: a
In this post we’ll demonstrate how to run deploy a simple scheduled task: a